Research
RESEARCH INTERESTS
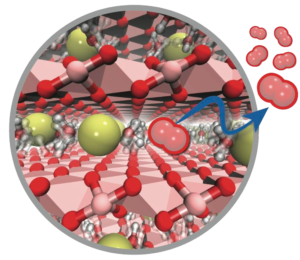
A portion of the materials science effort in the Klein group focuses on two-dimensional and layered materials for renewable energy technologies. Among their many applications, these materials are finding use as catalysts for renewable fuels, like water splitting and carbon dioxide reduction, and in photovoltaics and novel electronic devices. Our work in this area seeks to understand the fundamental physicochemical principles governing the use of layered materials in functional devices using a combination of quantum and statistical mechanics, computer simulations, and coarse-grained modeling techniques.
Our focus is on drug discovery, which encompasses a wide range of activities: learning the pathophysiology of a disease and using that data to develop a treatment that can stop or reverse its effects; testing chemical molecules biologically for potential therapeutic benefits against a wide range of disorders; discovering that repurposing existing pharmaceuticals in novel ways can have beneficial effects; and exploring novel treatment methods.
Biophysics is an ever-evolving interdisciplinary field that draws on the study of mathematics, chemistry, physics, engineering, pharmacology, and materials science to discover and create new methods for comprehending the intricate details of biology and, by extension, all life. We are working on ways to treat illness, end world hunger, generate clean energy, create innovative technology, and answer innumerable scientific puzzles.

Two-dimensional and Layered Materials for Renewable Energy Technologies
A portion of the materials science effort in the Klein group focuses on two-dimensional and layered materials for renewable energy technologies. Among their many applications, these materials are finding use as catalysts for renewable fuels, like water splitting and carbon dioxide reduction, and in photovoltaics and novel electronic devices. Our work in this area seeks to understand the fundamental physicochemical principles governing the use of layered materials in functional devices using a combination of quantum and statistical mechanics, computer simulations, and coarse-grained modeling techniques. This work is being done in close collaboration with experimentalists as part of Temple University’s Energy Frontier Research Center (EFRC) and the Center for the Computational Design of Functional Layered Materials (CCDM), and the Temple Materials Institute (TMI).
Anesthetics
Research on anesthetics at the ICMS focuses on the mechanisms by which volatile and intravenous general anesthetics modulate ion channels.
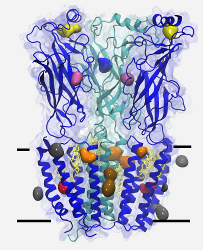
STRUCTURE OF TARGET RECEPTORS
nAChR: We have proposed that cholesterol (which is required for nicotinic acetylcholine receptor function) is deeply embedded in the transmembrane portion of the structure, and developed a model of a cholesterol-nAChR complex suitable for computational use. This model has been evaluated in simulations ranging up to 500 ns in length, and shown to significantly stabilize the receptor with respect to the control system.
K2P: We have built a model for the anesthetic-sensitive tandem pore domain potassium channel TREK-1, including the cytoplasmic domain. Although the cytoplasmic domain is initially constructed as a dimer extending into the cytoplasmic region, multiple simulations demonstrate that it docks as two monomers to the intracellular leaflet of charged lipid bilayers.
ANESTHETIC FORCE-FIELD DEVELOPMENT
Recently, CHARMM-compatible models for isoflurane and propofol have been developed within our group. Isoflurane parameters are available here, while propofol parameters are available upon request. Both models reproduce correct solvation free energies in TIP3P solvent.
FLOODING OF RECEPTORS BY ANESTHETIC
In this method, a high concentration of the anesthetic is placed in the aqueous phase of the simulation and allowed to partition into membrane and protein binding sites until a clinical concentration in water is reached. We have very recently provided detailed visual images of binding sites for isoflurane on nAChR and the homologous prokaryotic channel GLIC, demonstrating that the primary method of inhibition by isoflurane in these two channels is likely via pore-block. We are presently investigating the effect of isoflurane bound to potentially allosteric sites on nAChR dynamics.
REFERENCES:
- A. F. Barber, V. Carnevale, S. J. Raju, C. Amaral, W. Treptow, M. L. Klein, BBA-Biomembranes, 1818, 2120-2125 (2012). (Link)
- D. N. LeBard, J. Henin, R. G. Eckenhoff, M. L. Klein, G. Brannigan, PLOS Comput. Biol., 8 (5), e1002532 (2012). (Link)
- G. Brannigan, D. N. LeBard, J. Hénin, R. G. Eckenhoff, M. L. Klein, Proc. Natl. Acad. Sci. USA , 107(32), 14122-14127 (2010). (Link)
- W. Treptow, M. L. Klein, J. Am. Chem. Soc., 132, 8145-8151 (2010). (Link)
- J. Hénin, G. Brannigan, W. Dailey, R. Eckenhoff, M. L. Klein, J. Phys. Chem. B, 114, 604-612 (2010). (Link)
- Satyavani Vemparala, Carmen Domene, Michael L. Klein, Accounts of Chemical Research43, 103-110 (2010) (Link).
- L. Vedula, G. Brannigan, N. Economou, J. Xi, M. Hall, R. Liu, M. Rossi, W. Dailey, K. Grasty, M. L. Klein, R. Eckenhoff, P. Loll, J. Biol. Chem. 284, 24176-24184, (2009). (Link)
- C. A. Butts, J. Xi, G. Brannigan, A. A. Saad, S. P. Venkatachalan, R. A. Pearce, M. L. Klein, R. G. Eckenhoff, I. J. Dmochowski, Proc. Natl. Acad. Sci. USA 106, 6501-6506 (2009), (Link)
- G. Brannigan, J. Hénin, R. Law, R. Eckenhoff, M. L. Klein, Proc. Natl. Acad. Sci. USA 105, 14418-14423 (2008). (Link)
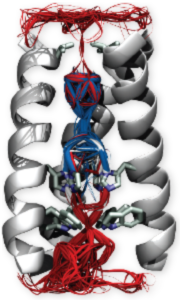
Voltage-gated cation channels
Voltage-gated cation channels are ubiquitous transmembrane proteins whose primary role is to transport selectively ions in a manner that depends on the polarization state of the membrane. They are involved in a variety of biological processes, among which the initiation and propagation of the nervous impulse. Mutations in genes encoding them are involved in a large number of inherited diseases (long QT syndrome, epilepsies, skeletal muscle paralyses, diabetes, deafness…). Voltage gated cation channels are composed of two main domains: the pore (including the gate, which opens and closes to let ions through or not, and the selectivity filter, which selects a specific type of ions) and the voltage sensor domain, which converts the electrical stimulus (change in the membrane potential) in a mechanical response (opening and closing of the channel).
CONDUCTION MECHANISM OF SODIUM
Selectivity is a fascinating propety of ion channels and can be attributed to the unique attributes of their selectivity filters. The resolution of the crystal structure of the first sodium selective channel has provided us with the first occasion to study the conduction and selectivity mechanisms in a such a selectivity filter. We use a combination of sophisticated simulation schemes (classical and QM/MM molecular dynamics simulation, free energy methods…) to compare these filters with the well-known potassium-selective ones.
FUNCTION AND MODULATION OF VOLTAGE-SENSOR DOMAINS
Voltage-sensor domains (VSD) are 4-helix transmembrane (TM) regulatory domains that undergo a conformational change in response to a change in the TM potential. Our work focuses on revealing the molecular-level fuctional mechanism of such domains and on providing an insight in their dysfunction due to genetic mutations.
REFERENCES:
- L. Stock, L. Delemotte, V. Carnevale, W. Treptow and M. L. Klein, J. Phys. Chem. B, 117, 3782-3789 (2013) (Link).
- L. Delemotte, M. L. Klein, M. Tarek, Front. Pharmacol., 3, 97 (2012) (Link)
- C. Amaral, V. Carnevale, M. L. Klein and W. Treptow, Proc. Natl. Acad. Sci., 109, 21336-21341 (2012) (Link).
- P. Gosselin-Badaroudine, L. Delemotte, A. Moreau, M. L. Klein, and M. Chahine, Proc. Natl. Acad. Sci., 109, 19250-19255 (2012) (Link).
- W. Treptow, M. L. Klein, J. Phys. Chem. Lett., 3, 1017-1023 (2012) (Link).
- E. Vargas, V. Yarov-Yarovoy, F. Khalili-Araghi, W. A. Catterall, M. L. Klein, M. Tarek, E. Lindahl, K. Schulten, E. Perozo, F. Bezanilla and B. Roux, J. Gen. Physiol., 140, 587-594 (2012) (Link).
- L. Delemotte, M. Tarek, M. L. Klein, C. Amaral, and W. Treptow, Proc. Natl. Acad. Sci., 115, 6109-6114 (2011) (Link).
- V. Carnevale; W. Treptow; M. L. Klein, J. Phys. Chem. Lett., 2 (19), 2504-2508 (2011) (Link).
- L. Delemotte, W. Treptow, M. L. Klein, M. Tarek, Biophys. J., 99, L72-L74 (2010) (Link).
M2 Proton Channel of Influenza Virus A
The M2 protein from influenza virus A is a proton channel, responsible for the acidification of the interior of the virion. This process is required for the release of the genetic material into the host cell. Its inhibition constitutes one of the major strategies for treating influenza, and as such it is the target of two of the four available flu drugs. Unfortunately the recent emergence of drug-resistant strains has neutralized these two drugs. Despite the fact that the recent determination of the channel structure has greatly increased our understanding of the function of M2, much of molecular mechanism of proton transport and of drug-resistance remains to be elucidated.
PROTON CONDUCTION MECHANISM
A full treatment of the dynamics of M2 is intrinsically challenging, however, because important events in the transport cycle occur on very different timescales and require different levels of description to model them accurately. Fast proton transfer processes are intrinsically coupled to much slower motions which are responsible for conformational changes in the protein. We adopt a combined approach involving experimental biophysics and theoretical investigations based on classical and hybrid quantum mechanics/molecular mechanics (QM/MM) molecular dynamics simulations. Our work elucidates the mechanism for proton transport in M2, where the protonation of specific pore-lining residues results in structural changes in the protein and pore waters.
DRUG BINDING
The design of effective anti-flu drugs requires a thorough understanding of the molecular mechanisms governing protein-drug binding and, more importantly, drug-resistance. The lack of structural information on the protein-drug complex, in particular for the S31N drug-resistant mutant, makes MD simulations a suitable approach to study the equilibrium configuration of channel-binding compounds. In order to capture the physico-chemical features impacting on the binding capabilities of a drug molecule, we use classical molecular dynamics simulations, complemented with approaches able to increase the sampling of rare events such as ABF and metadynamics, to screen libraries of compounds of known affinity.
REFERENCES:
- J. Wang, C. Ma, J. Wang, H. Jo, B. Canturk, G. Fiorin, L. H. Pinto, R. A. Lamb, M. L. Klein and W. F. DeGrado, J. Med. Chem., 56, 2804-2812 (2013) (Link).
- A. Bankura, M. L. Klein and V. Carnevale, Chem. Phys., in press (2013) (Link).
- J. Wang, Y. Wu, C. Ma, G. Fiorin, J. Wang, L. H. Pinto, R. A. Lamb, M. L. Klein and W. F. DeGrado, Proc. Natl. Acad. Sci., 110, 1315-1320 (2013) (Link).
- J. Wang, C. Ma, G. Fiorin, V. Carnevale, T. Wang, F. Hu, R. A. Lamb, L. H. Pinto, M. Hong, M. L. Klein, W. F. DeGrado J. Am. Chem. Soc., 133(32), 12834-12841 (2011) (Link).
- E. Khurana, R. H. DeVane, M. Dal Peraro, M. L. Klein BBA-Biomembranes, 1808, 530-537 (2011) (Link).
- V. Balannik, V. Carnevale, G. Fiorin, B. G. Levine, R. A. Lamb, M. L. Klein, W. F. DeGrado, and L. H. Pinto, Biochemistry, 49, 696-708 (2010). (Link)
- V. Carnevale, G. Fiorin, B. G. Levine, W. F. DeGrado, and M. L. Klein, J. Phys. Chem. C, 114 (48), 20856-20863 (2010) (Link).
- R. Acharya, V. Carnevale, G. Fiorin, B. G. Levine, A. L. Polishchuk, V. Balannik, I. Samish, R. A. Lamb, L. H. Pinto, W. F. DeGrado, M. L. Klein, Proc. Natl. Acad. Sci. USA,107,15075 (2010) (Link).
- E. Khurana, M. Dal Peraro, R. DeVane, S. Vemparala, W.F. DeGrado, M.L. Klein, Proc. Natl. Acad. Sci. USA, 106, 1069-1074 (2009). (Link).
Metalloenzymes
Metalloproteins constitute between one third and half of the proteome of living organisms and around 40% of the proteins deposited in the Protein Data Bank contain metals. Among many essential structural and biological functions, they are crucial in respiration and photosynthesis, participate in DNA and RNA processing and are involved in pharmacologically important reactions, such as metabolism of xenobiotics, antibiotic resistance or defense against reactive oxygen species.
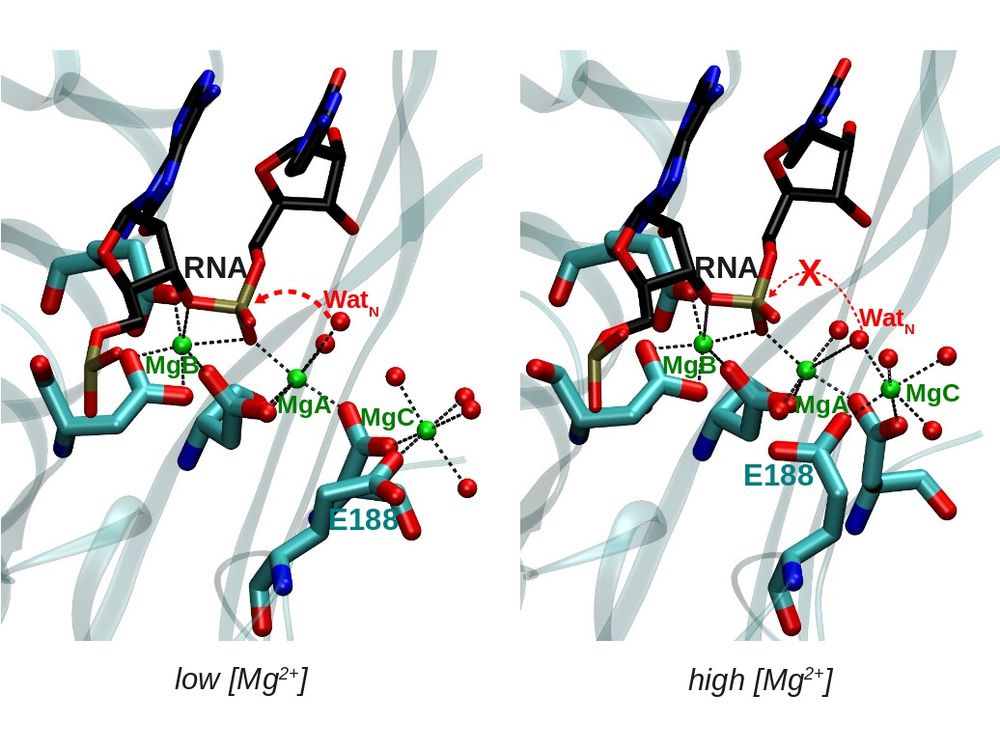
ENDONUCLEASES
Endonucleases are enzymes that cleave a phosphodiester bond within a RNA or DNA strand. The reaction is usually catalyzed by one or two divalent metal ions (in most cases Mg, but also Mn) bound to the enzyme. However, the number and nature of the metal ions involved in catalysis is not clear. Moreover, it is not well understood how the endonuclease activity is modulated by the metal concentration by post-translational modifications or by interaction with other proteins.
We use a combination of different computational methods (docking, classical molecular dynamics and QM and QM/MM calculations) in order to get further insight into the endonuclease activity of different ribonucleases.
This work is done in collaboration with the group of Prof. Allen W. Nicholson in the Department of Biology at Temple University.
REFERENCES:
- S. Xiao, M. L. Klein, D. N. LeBard, B. G. Levine, H. Liang, C. M. MacDermaid and M. Alfonso-Prieto, J. Phys. Chem. B, 118, 873-889 (2014). (Link)
- M. Alfonso-Prieto and M. L. Klein. “Density Functional Theory-based treatments of metal binding sites in metalloenzymes: challenges and opportunities” in Metalloproteins: structure, function and interactions, CRC Press, in press (2014). (Link)
- M.H. Ho, M. De Vivo, M. Dal Peraro, and M. L. Klein, J. Am. Chem. Soc., 132, 13702-13712 (2010). (Link)
Fusion of Biological Membranes
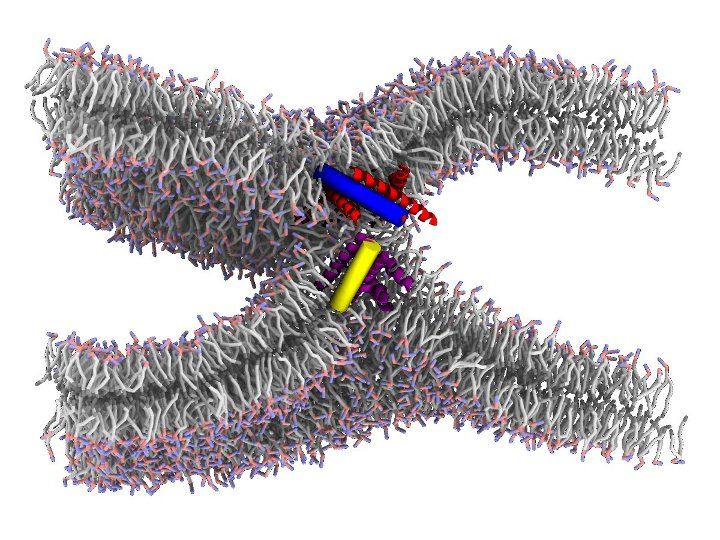
Membrane fusion is ubiquitous in biology: cell growth, extracellular transport and a multitude of processes involve membrane fusion. Fusion of synaptic vesicles to the plasma membrane is a key step in signal transmission between nerve cells. Within a cell, membrane budding and fusion enable non-disruptive transport of molecules between different organelles. Finally, viruses responsible for human disease, such as influenza and HIV, also begin the infection cycle by fusing their own membrane with the host cell’s.
The physical mechanism of membrane fusion, wherein two lipid bilayers merge to form a continuous structure, is typically unfavorable and influenced by factors including lipid composition, hydration and electrostatics. However, in addition to lipids biological membranes contain embedded proteins, often with sizable extracellular footprints that can interact with other proteins or with the opposing membrane. Such fusion proteins are initially bound to one of the two fusing membranes via a transmembrane helix domain, and utilize energetically favorable conformational transitions to lower the activation energy for membrane fusion.
It is well understood that fusion proteins shape the initial stages of the process, by facilitating the apposition between bilayers, and their dehydration as they come into more intimate contact. However, a major open question concerns the specific roles played by fusion proteins in the later stages of fusion. The “real” situation is likely to be intermediate between two hypothetical extreme scenarios: in the “lipid-centric” scenario, fusion proteins hold bilayers in close proximity, but remaining outside the point of membrane apposition, which is instead made up exclusively of lipids; in the “protein-centric” scenario, fusion proceeds via a pore lined with proteins, which remain at the very center of the point of contact as their conformational changes progress toward more stable states.
We are using simulations, all-atoms and coarse grained, combined with advanced sampling techniques, to test both scenarios on realistic examples of fusion. Our goal is to obtain a predictive computational model, to be used in understanding fundamental processes such as the transport of neurotransmitters, the design of new delivery systems and the infection by viruses.
References:
- J. E. Donald, Y. Zhang, G. Fiorin, V. Carnevale, D. R. Slochower, F. Gai, M. L. Klein, W. F. DeGrado Proc. Natl. Acad. Sci., 108(10), 3958-3963 (2011) (Link)
Translocation of Carbon Nanospheres Through Lipid Membranes
Carbon nanospheres (CNS) can be used for pharmaceutical and biomedical applications, such as drug delivery or cell imaging. It has been observed that such objects can enter into the nucleus of cells without any specific surface active chemical molecules. On the other hand there is the question whether such particles are toxic for human cells.

The translocation of carbon nanospheres through cell membranes is not fully understood on molecular scale, specifically nano-objects of sizes that are close or larger than the thickness of typical lipid membranes (around 3nm). In this study we investigate the mechanism of translocation of moderatly hydrophobic compounds through membranes without attaching any functional molecules on the translocated molecule. Fullerenes of different sizes serve as model system. We are specifically interested in the free energy change when a CNS enters the membrane.
We use all-atom Molecular Dynamics (MD) simulations to address this question. For CNS with sizes exceeding the bilayer thickness we develop a coarse grain model. Coarse grain simulations help to to overcome the required length and time scales, allowing for example to study also structures of CNS inside the membranes. The image shows a simulation snapshot of a C540 fullerene at the lipid-water interface.
References:
- A. Jusufi, R. H. DeVane, W. Shinoda, M. L. Klein, Soft Matter, 7, 1139 (2011). (Link)
Coarse Grain Models for Molecular Dynamics Simulations
Computational resources continue to rapidly increase allowing theoretical investigation of larger systems on longer timescales. Indeed the capacity of all-atom (AA) molecular dynamics (MD) has reached a level that permits somewhat routine exploration of systems containing on the order of hundreds of thousands of atoms for timescales approaching hundreds of nanoseconds. Nonetheless, with these spatial and temporal scales, many soft matter and biological systems of interest still extend far beyond this capability.


In order to overcome this issue, techniques such as enhanced sampling methods or reduced description models can be used, just to name a couple. Models using a reduced description of the system are typically referred to as coarse grain (CG) models. Recently there has been a renewed interest in CG techniques with numerous models now appearing in the literature. As well, there are numerous methods for deriving parameters for CG models.
However, each method has its limitations, which has limited greater adoption of these methods. One rather consistent characteristic of CG models is their dependence on AA MD simulations. This has the negative consequence of including any undesirable characteristics of the AA model into the CG model. We have recently developed a novel parameterization approach that relies heavily on experimental data including surface tension, density and free energy and reduces the dependence on atomistic molecular dynamics simulations. The resulting CG potential is based upon rather standard functional forms facilitating implementation in conventional MD codes. This approach has been applied to nonionic and anionic surfactants, biologically relevent lipid molecules and amino acids. The results demonstrate the ability to make modular transferable CG sites that are capable of accurately predicting the phase and surface behavior specific to a system.
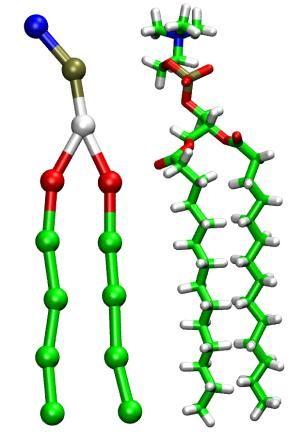
The image on the left shows the CG (left) and AA (right) representations of a DMPC lipid. The images on the right show the transition from a hexagonal phase to a lamellar phase of a PEG-lipid/water mixture upon reduction of the water content. Both are snapshots from simulations containing >1 mio. CG-beads.
CG models have been developed for non-ionic liquids which includes subsets of parameters for alkane, alcohol and ether based molecules. [1,2] Details of the water model can be found in this work as well. Our PC and PE based lipid model is available with flexibility in the hydrophobic chain length and degree of saturation. [3] Amino acids are currently in the development stage with some preliminary work reported. [4] We hope to bring the model up to speed soon. A very accurate phenyl based model with applications to fullerenes is available and has been integrated into our PC lipid model. [5,6,7] Ionic surfactants LABS [8] and SDS [9] along with counterions are also available. A more elaborate analysis of water models (not only including our current model) can be found in [10].
The specific functional forms required for the non-bonded potential of the CMM coarse grain model have been so far implemented into the following Molecular Dynamics simulation software packages:
LAMMPS (homepage)
LAMMPS is used by our group for most coarse grained simulations, and features excellent parallel scaling and many features. You have to enable the user-cg-cmm package (included in the main distribution) to use the CG potentials.
HOOMD-blue (homepage)
The HOOMD-blue package is MD program that runs entirely on graphics processors (GPUs) using the CUDA toolkit from Nvidia with up to 60x the speed of a single CPU.
MPDyn (homepage)
MPDyn is the “development” implementation of the CG model. Contact the MPDyn developer, Wataru Shinoda, to get access to a version that fully support the model. MPDyn is very efficient for serial execution and when using a small number of MPI tasks.
Tools to prepare and set up coarse grain simulations from scratch and from existing all-atom restart configuration are under development and will be published here, as well as the corresponding parameter and topology databased files.
References:
- W. Shinoda, R.H. DeVane, M.L. Klein, Mol. Sim. 33, 27-36 (2007)
- W. Shinoda, R.H. DeVane, M.L. Klein, Soft Matter 4, 2454 (2008)
- W. Shinoda, R.H. DeVane, M.L. Klein, J. Phys. Chem. B, 114, 6836 (2010)
- R.H. DeVane, W. Shinoda, P.B. Moore, M.L. Klein, J. Chem. Theor. Comput. 5, 2115-2124 (2009)
- R.H. DeVane, M.L. Klein, C. Chiu, S.O. Nielsen, W. Shinoda, P.B. Moore, J. Phys. Chem. B, 114, 6386 (2010)
- C. Chiu, R.H. DeVane, M.L. Klein, W. Shinoda, P.B. Moore, S.O. Nielsen, J. Phys. Chem. B, 114, 6394 (2010)
- R.H. DeVane, A. Jusufi, W. Shinoda, C. Chiu, S.O. Nielsen, P.B. Moore, M.L. Klein, J. Phys. Chem. B, 114, 16364 (2010)
- X. He, W. Shinoda, R.H. DeVane, K.L. Anderson, M.L. Klein, Chem. Phys. Lett. 487, 71-76 (2010)
- W. Shinoda, R.H. DeVane, M. L. Klein. In preparation. (2011)
- X. He, W. Shinoda, R.H. DeVane, M. L. Klein, Mol. Phys., 108, 2007-2020 (2010)
Collective Variables Module
Collective variables module main page
To tackle with the increasing complexity of macromolecular simulations, many techniques to enhance sampling have been made available in the past, most of them making use of reaction coordinates, or collective variables. Yet, multidimensional reaction pathways and conformational ensembles are most often modeled by very simple and ad hocone-dimensional coordinates. Frequently this happens not because of methodological constraints, but rather because of too inflexible implementations within the MD programs used.
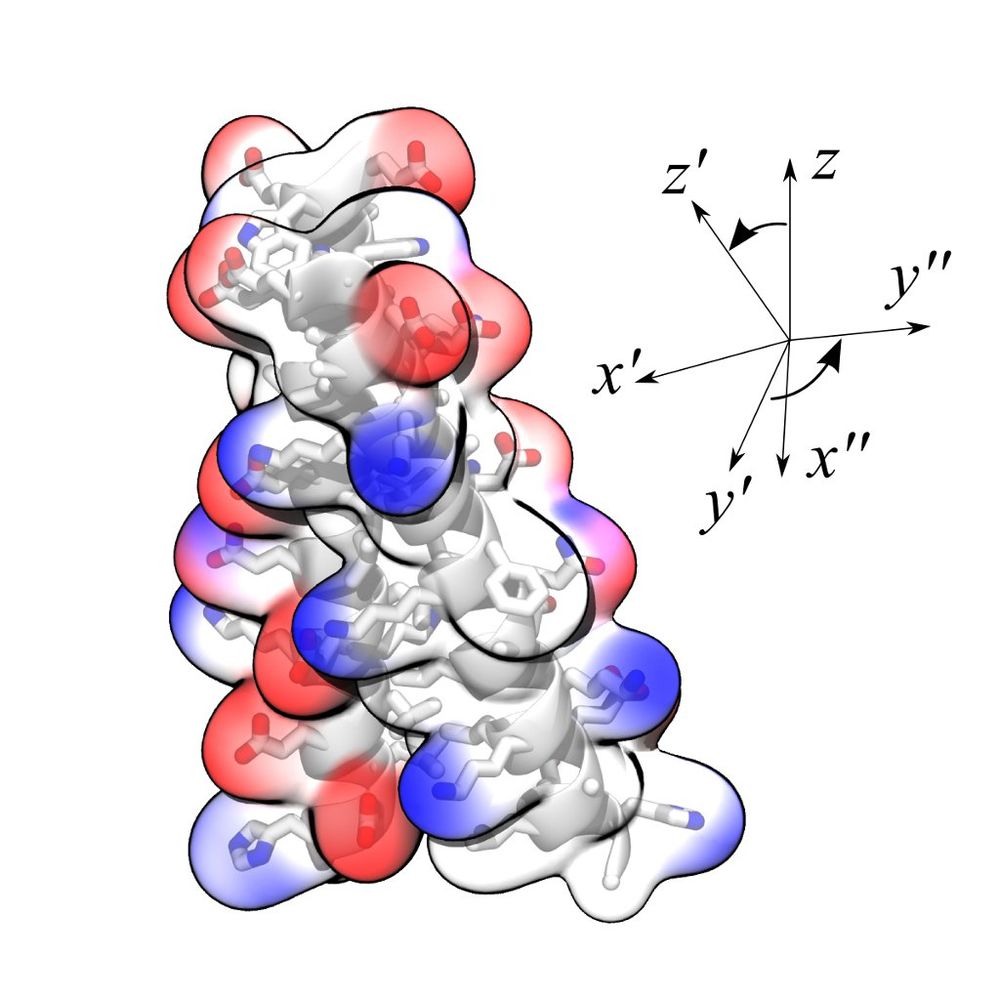
We have designed a module for MD codes that implements simultaneously several of such techniques, and an array of collective variables of broad usage. Currently available are: Adaptive Biasing Force (ABF), MetaDynamics (MtD), Steered MD (SMD) and Umbrella Sampling (US). There is no restriction on the number of variables, and many of their functional forms can be manipulated without recompilation. Rotational degrees of freedom are treated transparently, by generalizing the least-squares superposition approach of minimal RMSDs to model the orientations of macromolecules and flexible structures without artificial restraints. Statistical analyses can be performed at runtime, eliminating the need to store large trajectory files.
References:
- G. Fiorin, M. L. Klein and J. Hénin, Mol. Phys., in press (2013).
- J. Hénin, G. Fiorin, C. Chipot, and M. L. Klein, J. Chem. Theory Comput., 6, 35-47 (2010). (Link)
Contributed LAMMPS Packages
LAMMPS is a parallel molecular dynamics simulation software package with potentials for soft materials (biomolecules, polymers) and solid-state materials (metals, semiconductors) and coarse-grained or mesoscopic systems. It can be used to model atoms or, more generically, as a parallel particle simulator at the atomic, meso, or continuum scale. LAMMPS is designed to parallelize particularly well on large systems systems with no long-range interactions due to using a flexible spatial-decomposition of the simulation domain. The code provides a simulation framework that is easy to modify or extend with new functionality without losing parallel scaling.
Our contributions to LAMMPS include:
- fix imd for real-time visualization and interactive MD with VMD
- fix smd for steered MD
- pair_style cg/cmm etc. CMM coarse-grain pair potentials
- angle_style cg/cmm CMM coarse-grain angle/pair potential
Please see the Images and Movies Gallery page for some application examples.
Contributed VMD Plugins
VMD is a molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3-D graphics and built-in scripting. VMD supports computers running MacOS X, Unix, or Windows, is distributed free of charge, and includes source code. A member of the ICMS is a contributing VMD developer and also maintainting a number of VMD plugins listed below.
- TopoTools a Tcl script plugin library for manipulating topology information. It is meant to be a complementary tool to psfgen. TopoTools consists of a generic middleware script layer that provide convenient access to topology related data VMD. But it also has a number of high-level tools that allow reading and writing of topology file formats that cannot be parsed by molfile plugins, and replicating or combining multiple systems. TopoTools is bundled with VMD version 1.8.7 and later; updated versions may be found here.
- The signalproc plugin is a collection of scripted and compiled Tcl plugins that can be of use for signal processing and related types of applications within VMD.
- The gofrgui plugin provides a graphical user interface to measure gofr
- The irspecgui plugin is a GUI frontend to generating spectral densities.
- The clonerep plugin copies full sets of representations from one molecule to others.
- The dipwatch plugin allows to monitor the dipole moment of selections of atoms.
- The multimolanim allows to animate trajectories for systems with topology changes.
- CPMD trajectory file reader plugin
- X-Crysden xsf/axsf format reader
- HOOMD XML topology file reader.
Contributed HOOMD Modules
HOOMD-blue performs general purpose particle dynamics simulations on a single workstation, taking advantage of NVIDIA GPUs using the CUDA Toolkitto attain a level of performance equivalent a small cluster. Simulations are configured and run using simple python scripts, allowing complete control over the force field choice, integrator, all parameters, how many time steps are run, and more.
Our contributed modules to the HOOMD project for version 0.8.2 are:
PAST PROJECTS
Misdirected protein phosphorylation is frequently associated with human diseases, particularly cancer, neurological disorders, HIV, and diabetes. Over the last decade, there has been a growing interest in applying protein kinase inhibitors in cancer therapy. The first generation of protein kinase inhibitors use a traditional approach with a small drug molecule blocking the active site and is primarily targeting the cyclin dependend kinase 9 (CDK-9) activity. CDKs and their cyclin partners form a heterodimer, in which the CDK contains the enzymatic domain and the cyclin is the regulatory domain.
The goal of the project is to find more selective and specific ways to control CDK activity by regulating protein-protein binding instead. To that effect, novel drugs have to be designed on the basis of binding maps computed from all-atom and coarse grain molecular dynamics simulations.
References:
- V. Caracciolo, G. Laurenti, G. Romano, V. Carnevale, A. M. Cimini, C. Crozier-Fitzgerald, E. Gentile Warschauer, G. Russo, A. Giordano, Cell Cycle, 11, 1202-1216, (2012).
- G. Romano and A. Giordano, Cell Cycle, 7, 3664-3668, (2008).
Pathogenic bacteria contain two-component systems (TCS) that sense the environment within a host and activate mechanisms related to antimicrobial resistance.
Unraveling how binding of their ligands on the outside of the cell transmits a signal to the inside would help to understand the mechanisms of signal transduction.
This work is done in collaboration with Prof. William F. DeGrado (UCSF)
References:
- D. S. Goldberg, C. S. Soto, C. D. Waldburger, W. F. DeGrado., J. Mol. Biol., 379, 656-665 (2008).
- A. M. Stock, V. l. Robinson, P. N. Goudreau, Annu. Rev. Biochem., 69, 183-215 (2000).
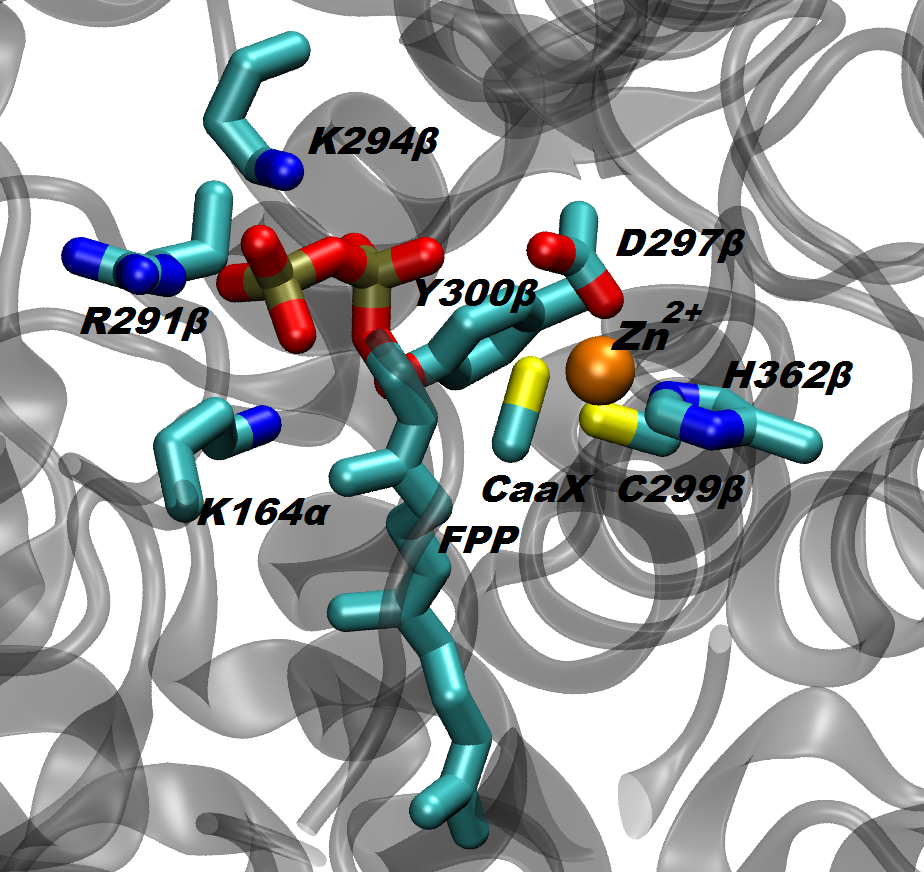
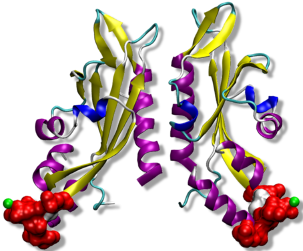
The protein farnesyltransferase (FTase), a Zn-metalloenzyme, catalyzes the transfer of the 15-carbon farnesyl group from the farnesyl diphosphate (FPP) to acceptor proteins that contain the so-called “CaaX” motif at the C-terminus. FTase activity is crucial in signal transduction pathways such as proliferation and apoptosis of cells. In fact, the Ras superfamily and small GTPases including Ras, Rho and Rab, are important examples of proteins that are activated by FTase function. Nowadays, FTase represents one of the promising targets for anti-cancer drug design, being involved in the activation of oncogene proteins such as mutated Ras, which are related to the development of ~20-30% of human cancers.
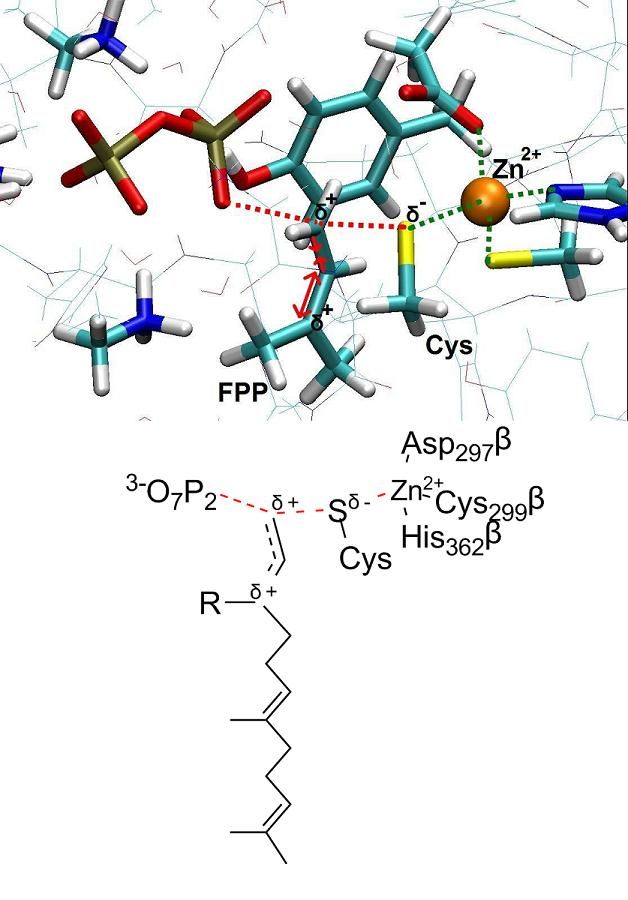
Several distinct hypotheses have been proposed based on various experimental findings in regard of the chemical reaction. A detailed picture of the reaction mechanism and its transition state would certainly improve our understanding of FTase enzymatic activity. Towards this aim, we focus on the chemical step of the catalytic cycle and present a theoretical investigation of the farnesylation mechanism. Our computations employ classical molecular dynamics (MD) and ab initio Car-Parrinello QM/MM calculations. Overall, our finding indicates that the reaction mechanism is an associative mechanism with dissociated character, which fits the experimental data well.
References:
- M.-H. Ho, M. De Vivo, M. Dal Peraro, and M. L. Klein, J. Chem. Theory Comput., 5, 1657 (2009)
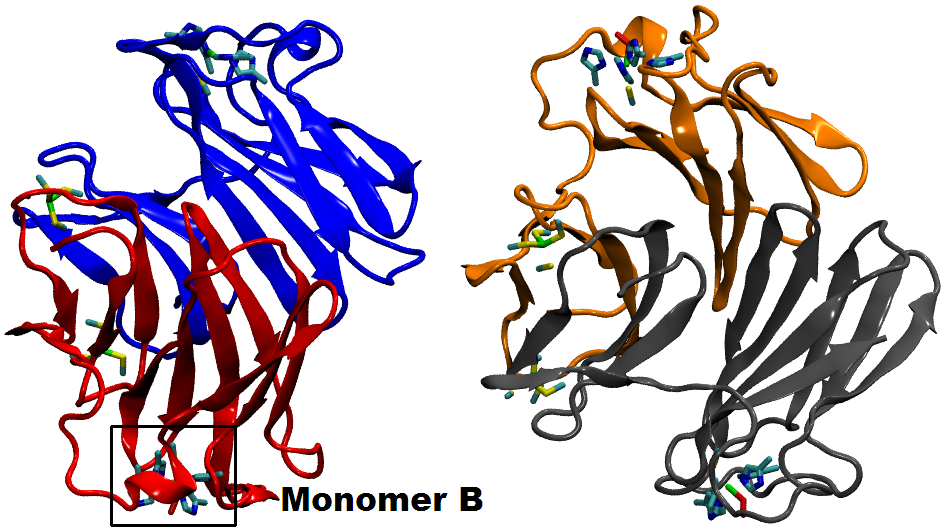
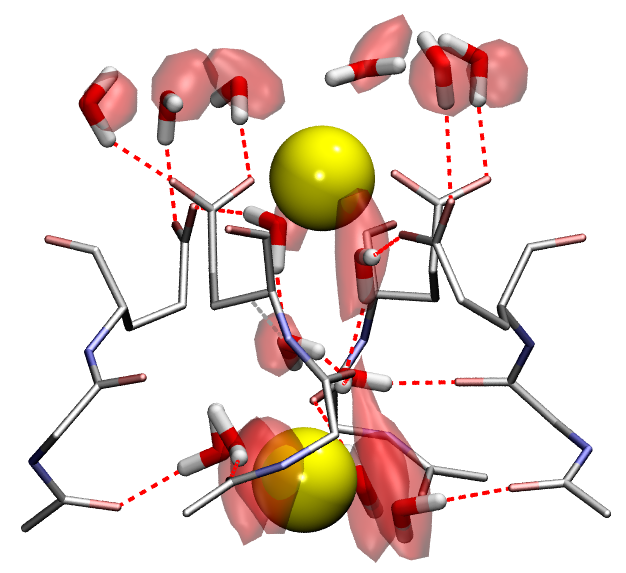
Many biological processes (including respiration, protein cleavage and toxic particles removal) are catalyzed by transition-metal containing enzymes. While localized d-states of the transition-metal ions are central in these catalytic reactions, accurate descriptions of these states face great challenges when using DFT method. In this work, we adopt a QM/MM technique coupled with a DFT + Hubbard U scheme in order to study the transition metal enzyme called Superoxide Reductase (SOR), with an appropriate description of the d-state electrons.
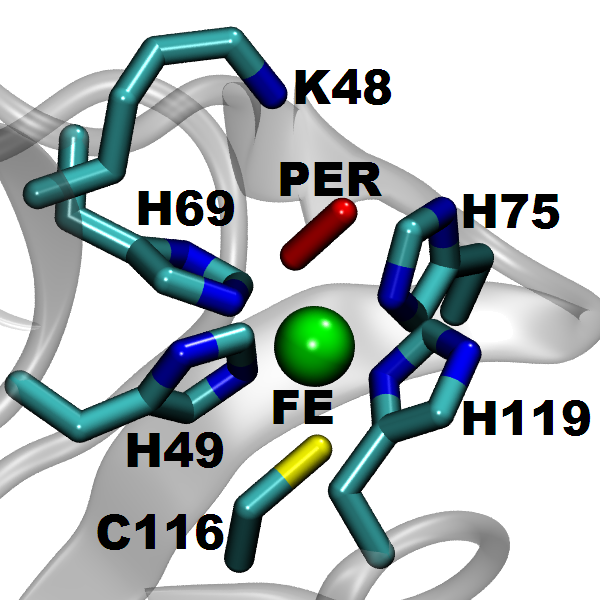
SOR (see the protein and active site structure shown in the figure), found in several anaerobic bacteria, is important in toxic oxygen derivatives removal, by catalyzing the one-electron reduction of superoxide to hydrogen peroxide. It prevents the production of molecular oxygen and opens a new way for the therapeutic treatment of diseases related to the presence of superoxide, through the ideation of suitable SOR mimics. We study the structure and energetics of the iron-dioxygen intermediates at different stages of the reduction process. According to our results, the introduction of the Hubbard U correction leads to an improved description of the intermediate structures and the reaction mechanism. Finite temperature studies with better descriptions of structures and energetics allow us to obtain a reliable quantitative analysis of the enzymatic reaction rate and mechanism.
References:
- P. H.-L. Sit, A. Migliore, M.-H. Ho and M. L. Klein, J. Chem. Theory Comput., 6, 2896-2909 (2010)
Electron-transfer reactions are ubiquitous processes in organic and inorganic redox reactions. In the diabatic limit, the transfer integral is small, and the system has to diffuse up to the intersection several times before an electron can tunnel. The reaction rate for such a weak coupling case can be written as
where Hif is the transfer integral between the initial and final states. ρFC is the density of states weighted by the Franck-Condon factor and thermally averaged. This term becomes
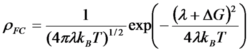
in the classical case. In the formula, λ is the reorganization energy and ΔG is the energy of the reaction.
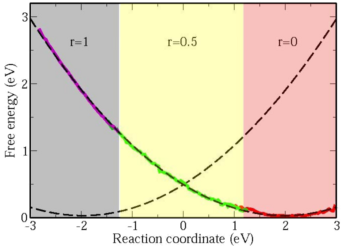
To obtain quantitative descriptions of electron-transfer reactions fully from first-principles, we developed a novel method to calculate free-energy surfaces of electron-transfer reactions from ab initio molecular dynamics. Sampling of phase space is done with a penalty functional added to the standard density functional so that the oxidation states of ions can be controlled. This penalty functional opens up the possibility of performing out of ground state DFT calculations, and is also a practical way to correct for the self-interaction in DFT.
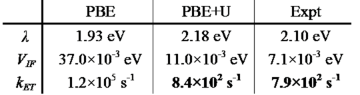
With this penalty functional technique, we calculated the free-energy surfaces of the self-exchange reaction between aqueous ferrous and ferric ions fully from first-princieples (see figure). We found that the free-energy curves are parabolic, validating the linear solvation model first proposed by Marcus. The reorganization energy obtained is 1.93 eV, which is in good agreement with the experimental value of 2.1 eV, and a big improvement over classical and semi-classical calculations in quantitatively describing electron-transfer reactions. A further improved value of 2.18 eV is obtained by including Hubbard U correction to accurately describe the strongly localized d-electrons.
We then calculated the transfer integral by adopting a recently developed ab initio method. Combining the reorganization energy and the transfer integral, we can estimate the ET reaction rate constant fully ab initio. The result of 8.4 × 102 s-1 is in excellent agreement with the experimental estimate of 7.9 × 102 s-1.
References:
- A. Migliore, P. H.-L. Sit, and M. L. Klein., J. Chem. Theory Comput., 5, 307 (2009)
- P. H.-L. Sit, M. Cococcioni, and N. Marzari. J. Electroanal. Chem., 607, 107 (2007)
- P. H.-L. Sit, M. Cococcioni, and N. Marzari. Phys. Rev. Lett., 97, 028303 (2006)
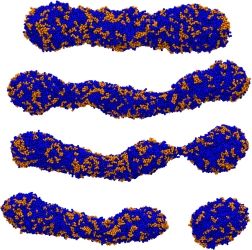
A worm-like micelle is a classic example of a molecular self-assembly that possesses a unique elongated shape. The polymer hydrophobic tails self assemble in the core of the micelle, while the hydrophilic groups maximize contacts with water by self-assembling in the corona of the micelle. Using mesoscopic simulation techniques, dissipative particle dynamics (DPD), we can test the stability of worm-like micelles at different hydrophilic block fractions. Worm-like micelles exist in a narrow region of the phase diagram. If the hydrophobic portion of the diblock copolymer is degradable, such as PCL (poly(caprolactone) (PCL) or poly(lactic acid) (PLA), this leads to shortening of the relative molecular weight of the tail, and an increase in the polydispersity, over time. The leads to instabilities, and ultimately, a breakup of the worm-like micelle morphology. We emulate this process in simulation by mixing in diblocks of differing hydrophobic length. At a critical concentration, this leads to a breakup of the previously stable worm. This instability begins with radial undulations along the core, bud formation at the end of the micelle, and finally expulsion of a spherical micelle from the end-cap. Analysis of the mean curvature of the worm micelle during break-up proves a simple linear relationship with the mean interfacial concentration of each component diblock copolymer relative to each equilibrium unmixed micellar phase.

Other examples of self-assembly structures are bilayers. We use Coarse-Grained Molecular Dynamics (CGMD) to gain insight about the strong lateral segregation observed in polymersomes composed by neutral and charged diblock-copolymers (Christian et al, 2009) in the presence of calcium. Our approximation involved toy models in which the charged groups were represented with LJ interaction sites. These simple models are able to successfully reproduce the morphology phase diagram for charged diblock copolymers as well as the spontaneous segregation observed in bilayers composed by charged and neutral copolymers. The shape of the aggregates appeared correlated to its state: crystalline patches are irregular, liquids are regular. As suggested by experiments, after several nanoseconds of simulation, we found strong correlation between the local composition of the two leaflets (registration). Simulations suggest that differentiated interlealet interaction between ordered and disordered phases would contribute to the registration.
References:
- S. M. Loverde, M. L. Klein and D. E. Discher, Advanced Materials, 24, 3823-3830 (2012)
- D. A. Pantano, P. B. Moore, M. L. Klein and D. E. Discher, Soft Matter, 7, 8182-8191 (2011)
- S. M. Loverde, V. Ortiz, R. D. Kamien, M. L. Klein and D. E. Discher, Soft Matter, 6, 1419-1425 (2010)
- D. A. Christian, A. Tian, W. G. Ellenbroek, I. Levental, K. Rajagopal, P. A. Janmey, A. J. Liu, T. Baumgart & D. E. Discher, Nature Materials 8, 843-849 (2009)
- Y. Geng and D. E. Discher.Journal of the American Chemical Society, 127, 12780-12781 (2005)
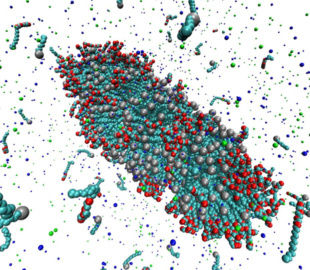
Surfactants are used in many industrial applications, such as detergents in cleaning products, as visco-elastic drilling fluids, or for solubilization of lipid molecules. Above the so-called critical micelle concentration (cmc), surfactants self-assemble into aggregates of characteristic morphologies (spherical or worm-like micelles, bilayers) depending, e.g., on the chemical architecture of the surfactant or the composition of the surfactant solution. It is computationally very challenging to model the self-assembly process of such systems, due to the required systems sizes and the long aggregation time (>1μs). Coarse grain simulations and implicit solvent models allow studies of self-assembly processes on the required time and length scales. A recently developed implicit solvent model of ionic surfactants is extended to study the effect of divalent salt ions (Ca2+ or Mg2+) on the micellization properties. Furthermore we investigate micellar properties when different surfactant types are mixed, such as sodium dodecyl sulfate (SDS) with polyethylene glycol (PEG) surfactants. It has been experimentally observed that the composition of the surfactant mixture and length of the PEG head group have significant impact on the morphologies of the micelles, e.g., the formation of spherical and worm-like micelles. We are particularly interested in such synergetic effects, and the ability to quantitatively predict the cmc and agggegate structures which is not feasible using conventional atomistic simulation approaches. We combine Grand Canonical Monte Carlo and atomistic Molecular Dynamics simulations to parameterize the model and to study the mentioned self-assembly properties. The image shows a simulation snapshot of an ellipsoidal micelle composed of a mixture of sodium dodecyl sulfate [SDS; grey spheres attached to light blue chain] and dodecyl polyethylene glycol surfactants [red and light blue chain molecules] in presence of NaCl salt [small dark blue and green particles].
References:
- A. Jusufi et al., J. Phys. Chem. B 112, 13783 (2008)
Meeting the demands of the rapidly advancing nanotechnological frontier requires novel, multifunctional nanoscale materials. Among the most promising nanomaterials to fulfill this need are biopolymer-carbon nanotube hybrids (Bio-CNT). Bio-CNT consists of a single-walled carbon nanotube (CNT) coated with a self-assembled layer of biopolymers such as DNA or protein. Experiments have demonstrated that these nanomaterials possess a wide range of technologically useful properties with applications in nanoelectronics, medicine, homeland security, environmental safety and microbiology. We use all-atom molecular dynamics (MD), parallel tempering replica exchange (REMD) and free energy methods gain insight into properties and behavior of these unique nanomaterials.
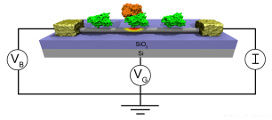
Detection of viruses and other harmful biological agents has important applications in homeland security and medicine. Recently, the experimental nanoscience groupof Dr. Charlie Johnson at the University of Pennsylvaniahas developed sensors capable of detecting proteins associated with the adenovirus (one of the viruses responsible for the common cold). These sensors are depicted below. They consist of the Coxsackie-adenovirus receptor protein (green) attached to a carbon nanotube (gray cylinder). Knob proteins (orange) that are located on the surface of the adenovirus bind to this receptor protein. This binding event changes the electrical properties of the carbon nanotube. Thus, by monitoring the electrical properties of the carbon nanotube, one can detect the presence of the adenovirus. ICMS uses a variety of computational techniques to help rationalize the fabrication and operation of these sensing devices.
References
- R.R. Johnson, A.T.C. Johnson, M. L. Klein, Small, 6, 31, (2010).
- R.R. Johnson, B.J. Rego, A.T.C. Johnson, M.L. Klein, J. Phys. Chem. B, 113, 11589 (2009)
- R.R. Johnson, A. Kohlmeyer, A.T.C. Johnson, M. L. Klein. Nano Lett., 9, 537 (2009)
Ionic liquids made up of organic cations and organic or inorganic anions have very low vapor pressure, which sets them apart from commonly used organic solvents. This makes them attractive for use as solvents for synthesis and catalysis and other industrial applications.
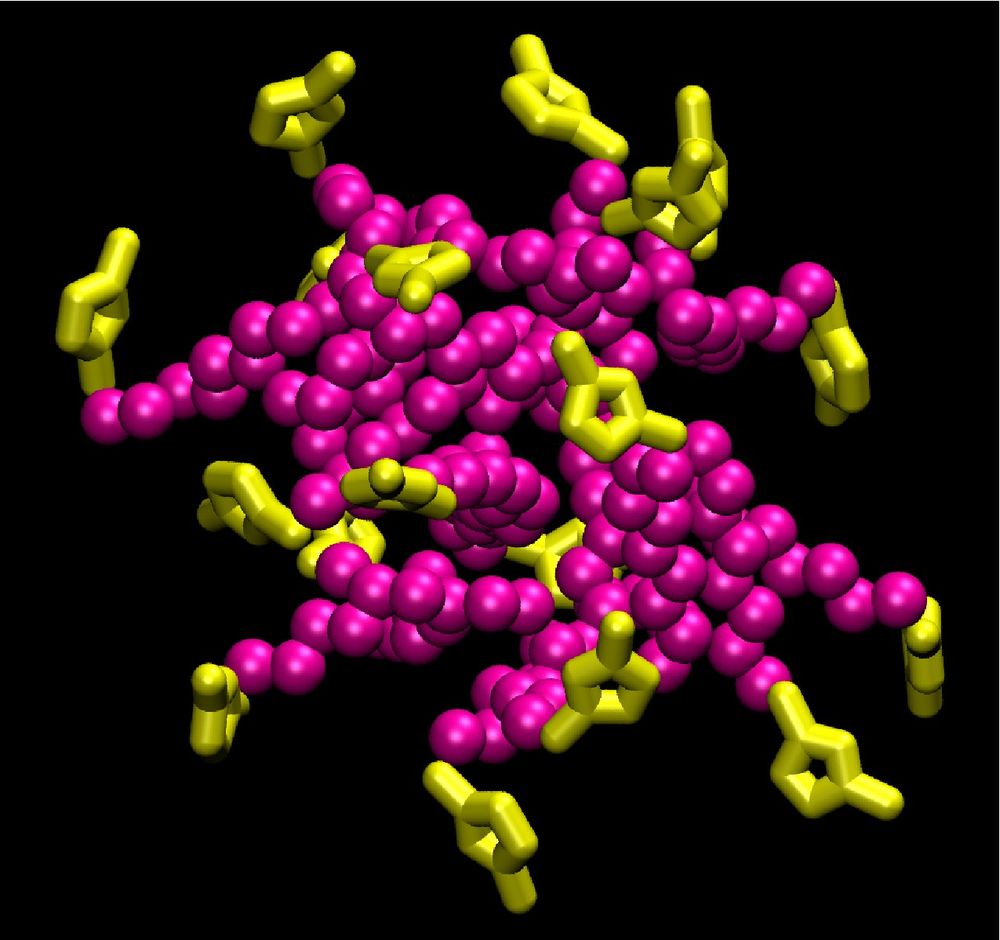
Atomistic molecular dynamics (MD) studies have been performed to study aggregation in these systems. Aqueous solutions of [C10mim][Br] spontaneously form cation aggregates. Simulations of the vapor-liquid interface of the same system shows that the alkyl tails of the cations aggregate at the surface to minimize the unfavorable interactions with water. In higher concentration, the cations also form aggregates in the aqueous phase with polar head groups (imidazolium ring) pointing outwards and the alkyl tails inwards. The determined aggregate sizes agree with experimental observations. The simulated systems appear to be in a metastable state with reference to the aggregation numbers. To extend the accessible observation times, a coarse grained model has been developed for this system.
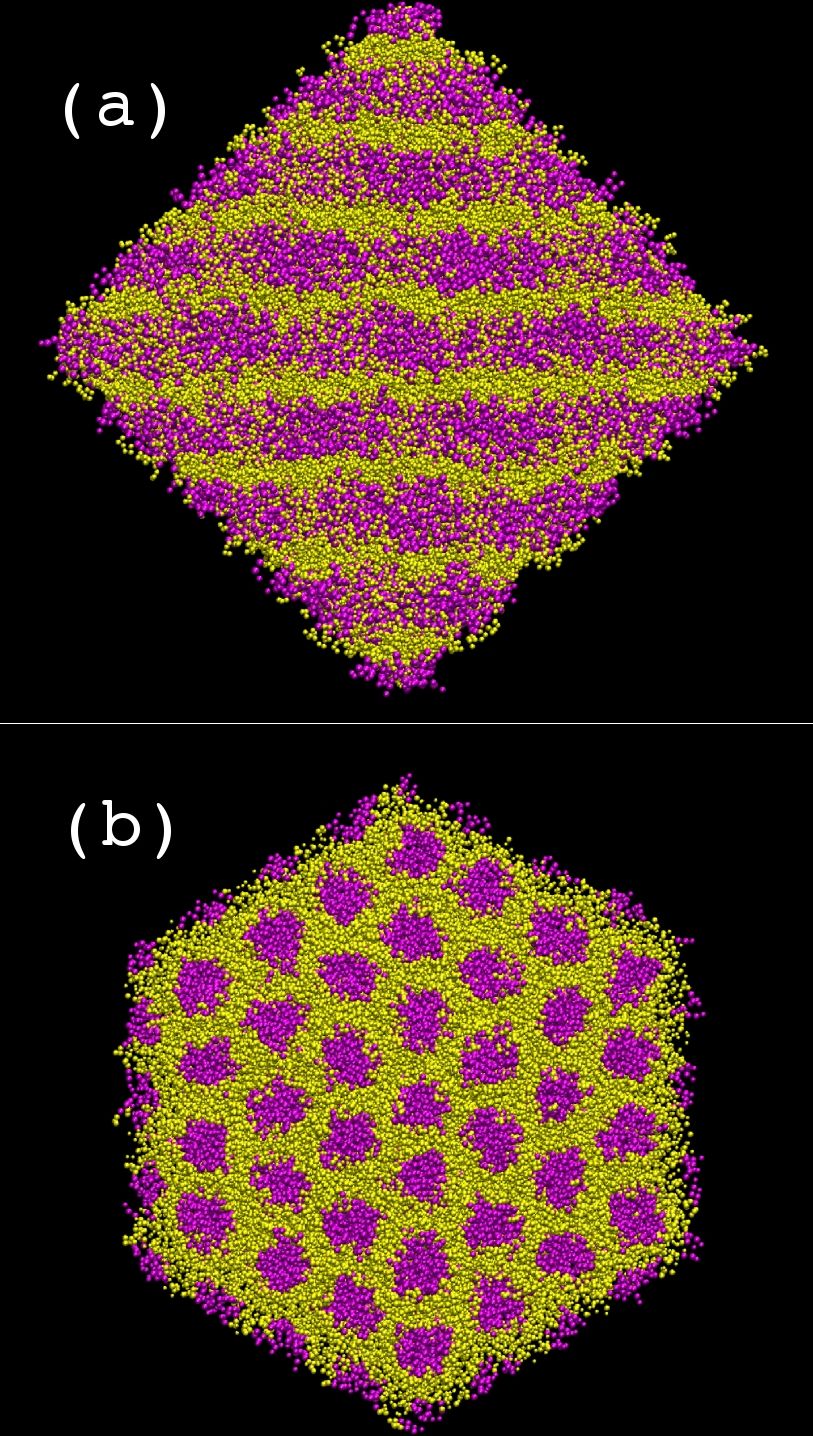
Coarse grain MD simulations of aqueous [C10mim][Br] solutions at several concentrations provide insight into their microscopic structure. At 37% (w/w) water concentration there is a separation of hydrophilic (head groups, anions and water, yellow) and hydrophobic regions (tail groups, magenta) forming a hexagonal columnar phase (see figure on the right: (a) side view of columns, (b) top view of columns).
In dilute aqueous solutions of [C10mim][Br] quasi-spherical micelles form spontaneously. Initially, a large number of monomers and few small aggregates are observed. Yet over time the aggregates grow in size by incorporating those monomers.
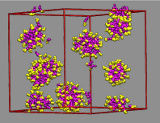
For the small aggregates it is difficult to grow by merging with other micelles due to repulsion of their charged surfaces. The observed aggregates are poly-disperse and typically contain between 40 and 60 cations. The bulk region of the vapor/liquid interface of aqueous ionic liquids shows similar aggregation behavior.
References:
- B.L. Bhargava, M. L. Klein, J. Phys. Chem. A., 113, 1888 (2009)
- B.L. Bhargava, M. L. Klein, Mol. Phys., 107, 393 (2009)
- B.L. Bhargava, M. L. Klein, Soft Matter, 5, 3475 (2009)
- B.L. Bhargava, M. L. Klein, J. Phys. Chem. B, 113, 9499 (2009)
